%LEAD_DISTANCES_LATEX Script to extract data on lead accuracy
%
% Provides data for latex reports.
%
% Usage: define target, STN, GPI, VIM
%
% Outputs: .mat & .csv file of distances + plots & coords
%
% NB: needs ea.stats open
% NNB: set for atlas Distal (medium)
%
% Michael Hart, University of British Columbia, August 2020
%LEAD_LOGBOOK Report on electrode accuracy (e.g. for a surgical logbook)
%
% Usage: define target, STN, GPI, VIM
% define "in" distance, distance considered inside the target
%
% Outputs: .mat & .csv file of distances + plots & coords
%
% NB: run from patient directory - needs ea.stats open
% NNB: all atlases are Distal (medium)
%
% Michael Hart, University of British Columbia, November 2020
%%lead_radar
%
% Script for analysing electrode targeting
%
% Just set group directory & target below
%
% NB: set for distal medium atlas
% NNB: saves & returns to group directory
%
% Michael Hart, University of British Columbia, December 2020



%LEAD_FLIPPER Duplicates leads for viewing single side results as a group
%
% Usage: subject to duplicate, absolute path
%
% Outputs: ea_reconstruction.mat in new lead_flipped folder within working directory
%
% NB: set for Medtronic 3389
%
% NNB: code based on discussion here
% [https://www.lead-dbs.org/forums/topic/export-code-for-vta-calculation/]
%
% Michael Hart, University of British Columbia, November 2020
%MULTI_VAT Compares difference VAT models
%
% Makes VATs with multiple (3-4) models
% Outputs VATs in separate folders
% Also DICE coefficient confusion matrix
%
% nb: works from patient directory
% nnb: need to run first stimulation in Gui & name it "vat_horn"
%
% Michael Hart, University of British Columbia, May 2021





=============================================================================================
tract_van.sh
(c) Michael Hart, University of British Columbia, August 2020
Co-developed with Dr Rafael Romero-Garcia, University of Cambridge
Function to run tractography on clinical DBS data (e.g. from UBC Functional Neurosurgery Programme)
Based on the following data: GE scanner, 3 Tesla, 32 Direction DTI protocol
Example:
tract_van.sh --T1=mprage.nii.gz --data=diffusion.nii.gz --bvecs=bvecs.txt --bvals=bvals.txt
Options:
Mandatory
--T1 structural (T1) image
--data diffusion data (e.g. standard = single B0 as first volume)
--bvecs bvecs file
--bvals bvals file
Optional
--acqparams acquisition parameters (custom values, for Eddy/TopUp, or leave acqparams.txt in basedir)
--index diffusion PE directions (custom values, for Eddy/TopUp, or leave index.txt in basedir)
--segmentation additional segmentation template (for segmentation: default is Yeo7)
--parcellation additional parcellation template (for connectomics: default is AAL90 cortical)
--nsamples number of samples for tractography (xtract, segmentation, connectome)
-d denoise: runs topup & eddy (see code for default acqparams/index parameters or enter custom as above)
-p parallel processing (slurm)*
-o overwrite
-h show this help
-v verbose
Pipeline
1. Baseline quality control
2. FSL_anat*
3. Freesurfer*
4. De-noising with topup & eddy - optional (see code)
5. FDT pipeline
6. BedPostX*
7. Registration
8. XTRACT (including custom DBS tracts)
9. Segmentation (probtrackx2)
10. Connectomics (probtrackx2)*
Version: 1.0
History: original
NB: requires Matlab, Freesurfer, FSL, ANTs, and set path to codedir
NNB: SGE / GPU acceleration - change eddy, bedpostx, probtrackx2, and XTRACT calls
=============================================================================================

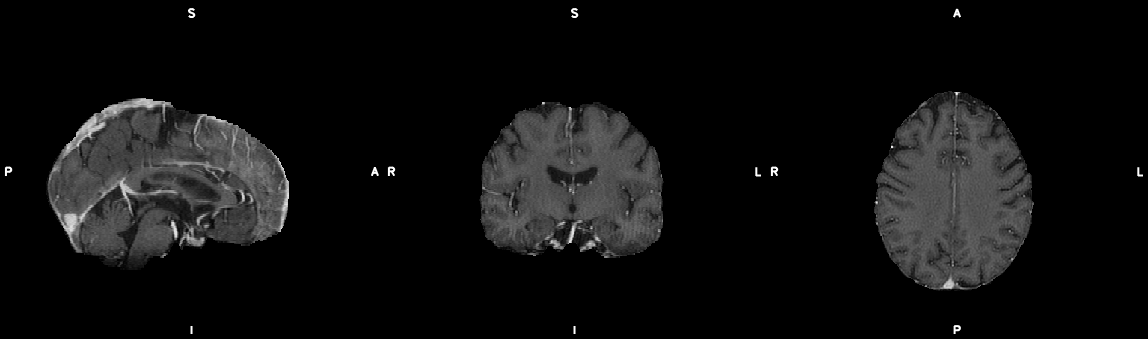
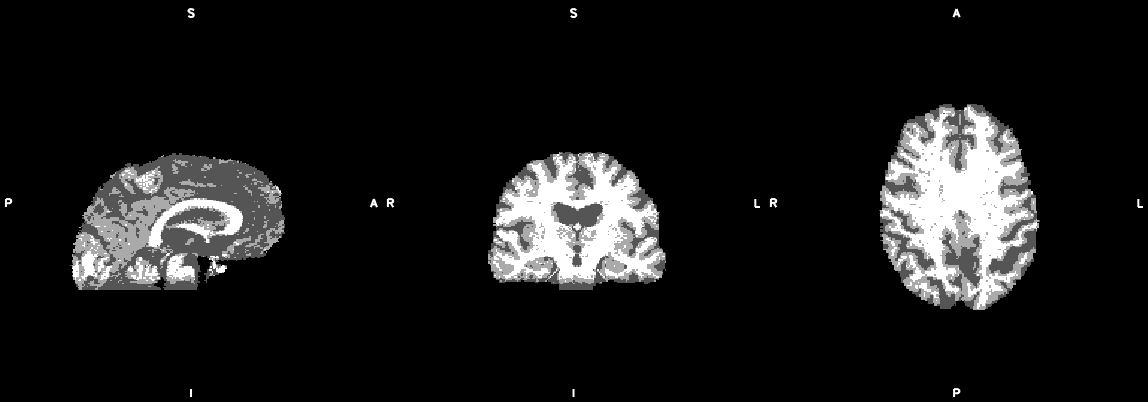
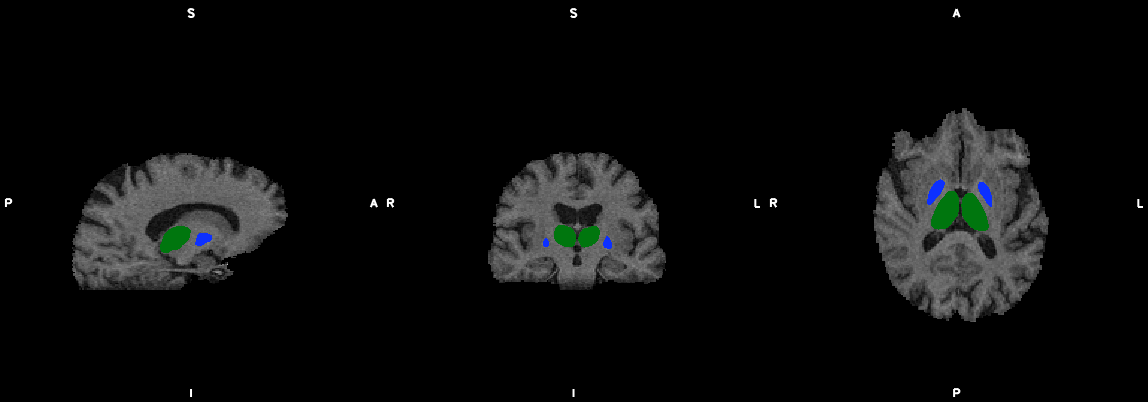
#1. Make single slice bedpostx files & submit to cluster
for ((slice=0; slice<${nSlices}; slice++)); #loop through all of the parts you wish to run in parallel
do
....
#make a bash file for your command
#this is the actual commands
echo 'bedpostx_single_slice.sh ${tempdir}/diffusion ${slice} --nf=3 --fudge=1 --bi=1000 --nj=1250 --se=25 \
--model=1 --cnonlinear' >> ${tempdir}/diffusion.bedpostX/command_files/command_`printf %04d ${slice}`.sh
#now submit it to the cluster via Slurm / sbatch
sbatch --time=02:00:00 ${tempdir}/diffusion.bedpostX/command_files/command_`printf %04d ${slice}`.sh
....
done
#2. Combines individual file outputs
#Check if all made: if not, resubmit for longer
....
if [[ "${bedpostFinished}" -ne "${nSlices}" ]] ; #check if the number of outputs matches the number of segments planned to run
then
for ((slice=0; slice<${nSlices}; slice++)); #loop through all segments above
do
echo ${slice}
if file_doesnt_exist
then
....
#resubmit bash file above for longer if doesn't exist
....
fi
done
#3. If all made, run bedpostx_postproc to combine
#This says it all, just run the same check as in step 2 then it's a one line command
fi
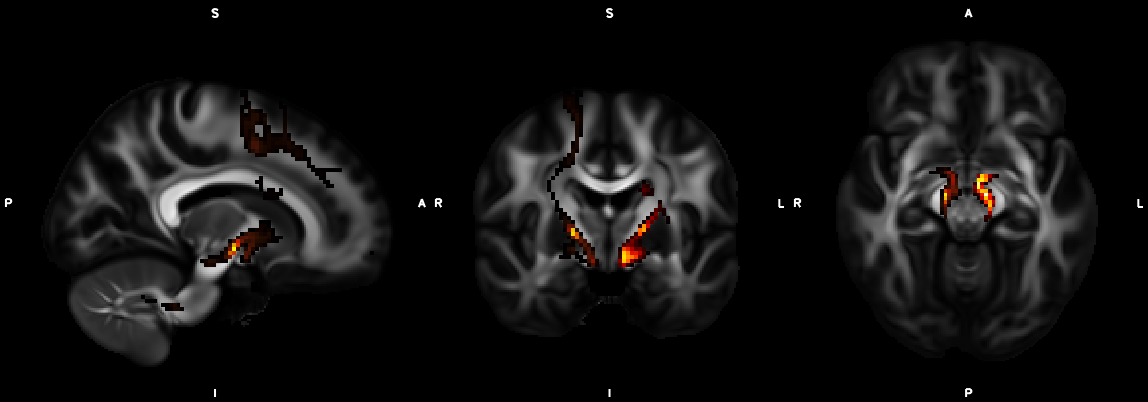
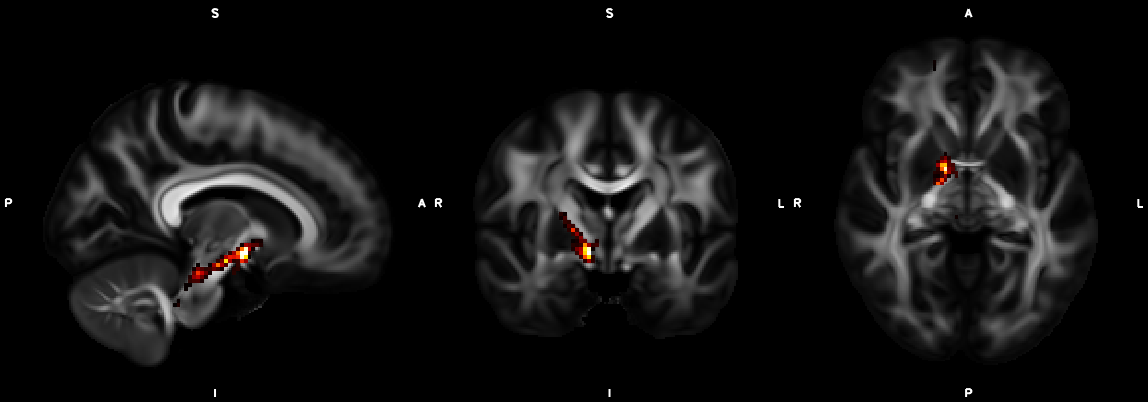
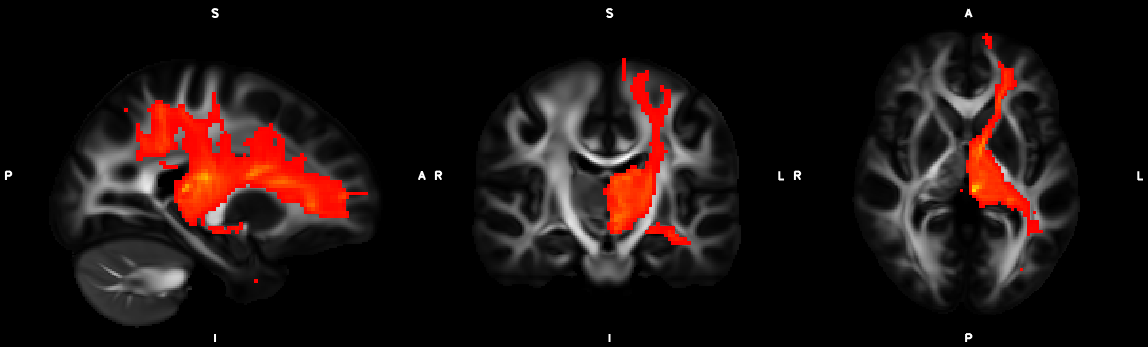
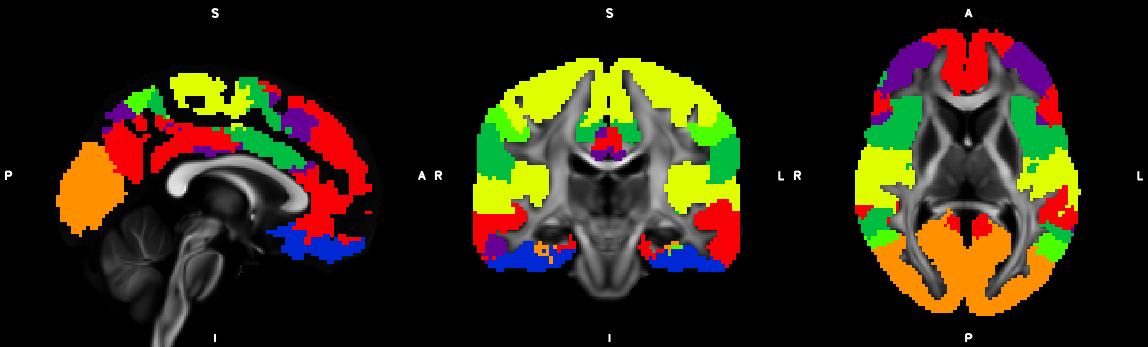
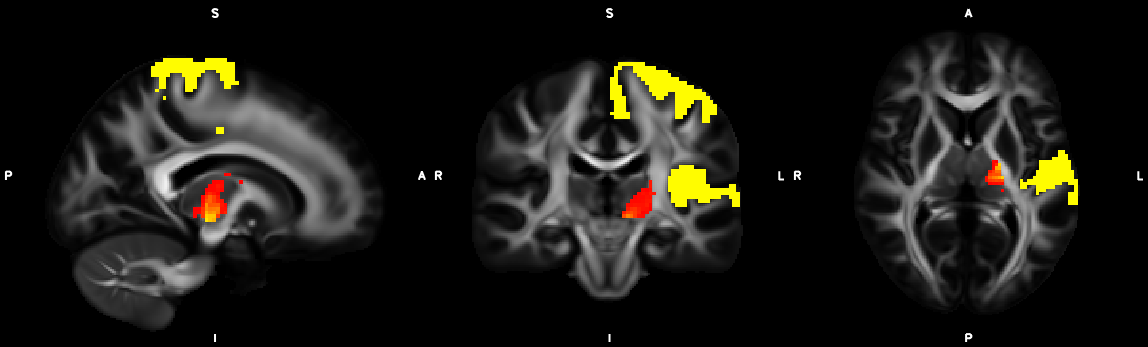
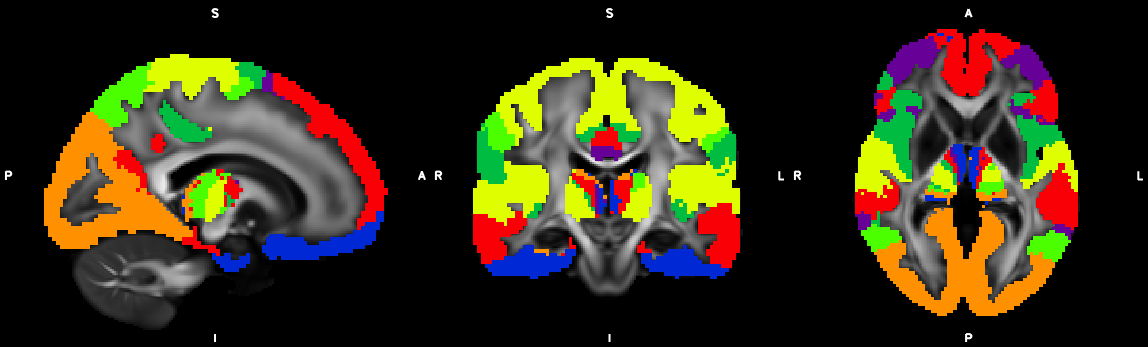
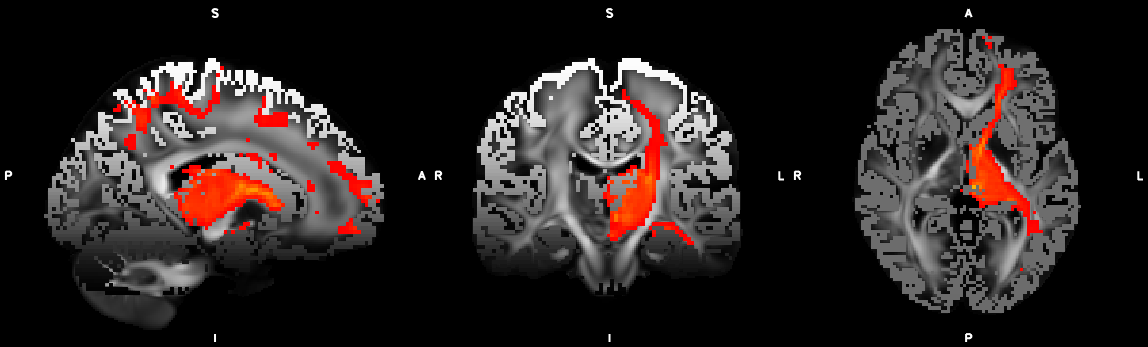
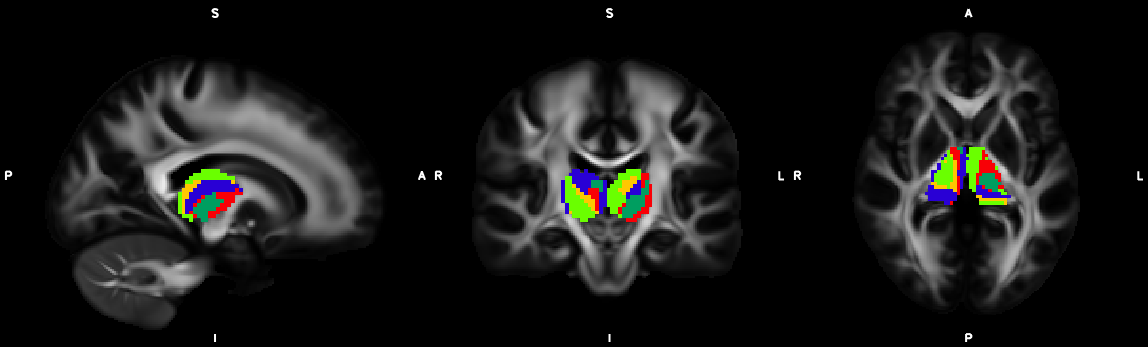
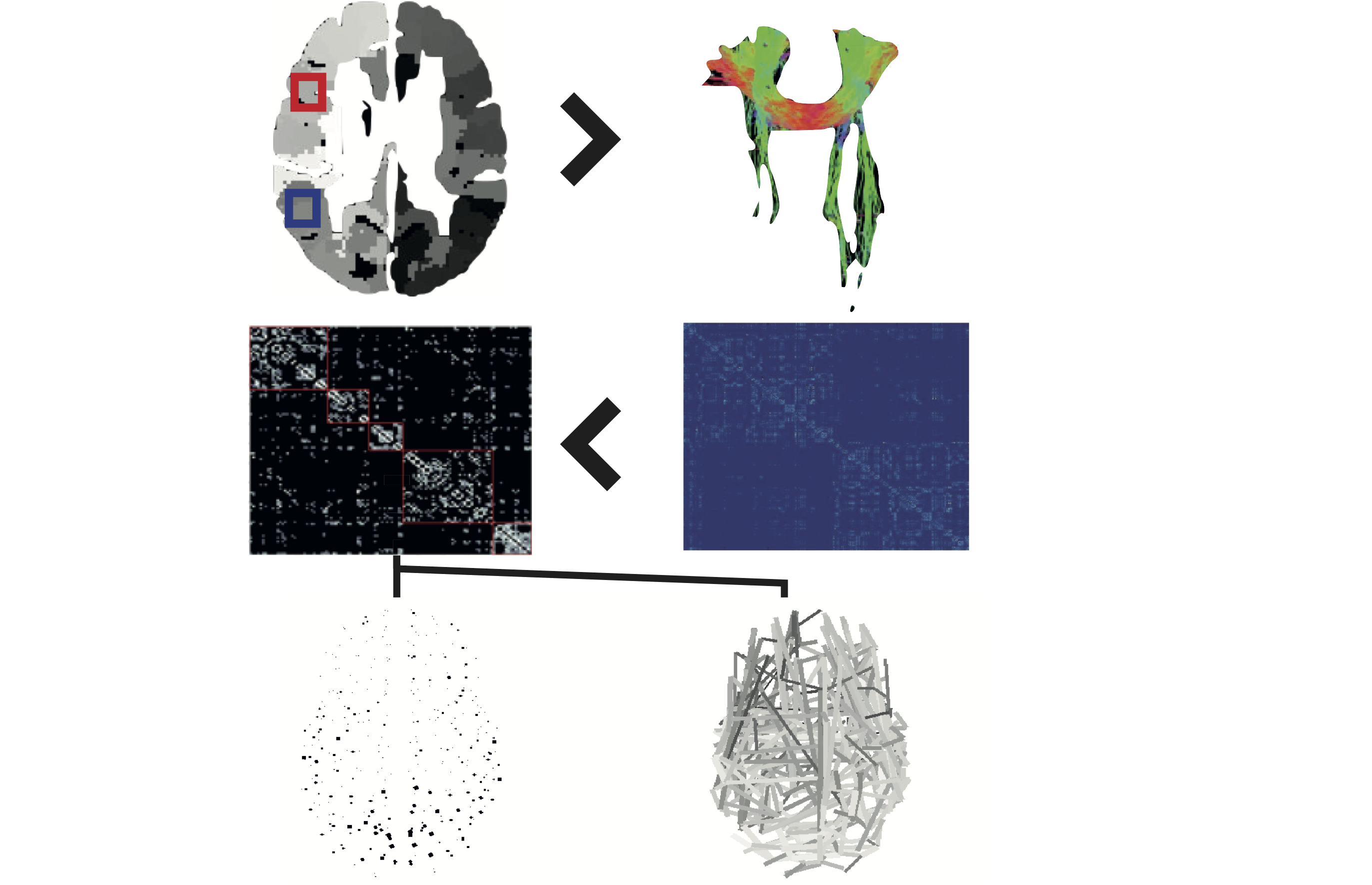
%% Script for connectome analysis with tractography data
%
% Dependencies: BCT 2019_03_03, contest, versatility, matlab_bgl, powerlaws_full, schemaball, BrainNetViewer
%
% Inputs: data.txt, connectivity streamlines matrix
% xyz.txt, parcellation template co-ordinates
%
% Outputs: graph theory measures & visualisations
%
% Version: 1.0
%
% Includes
%
% A: Quality control
% A1. Load data
% A2. Basic definitions
% A3. Connectivity checks
% A4. Generation of comparison graphs
%
% B: Network characterisation
%
% B1. Modularity & Versatility
% B2. Graph theory measures
% B3. Normalise measures
% B4. Measures statistics
% B5. Symmetry
% B6. Measures plotmatrix
% B7. Cost function analysis
% B8. Small world analysis
% B9. Degree distribution fitting
%
% C: Advanced network topology
%
% C1. Hubs
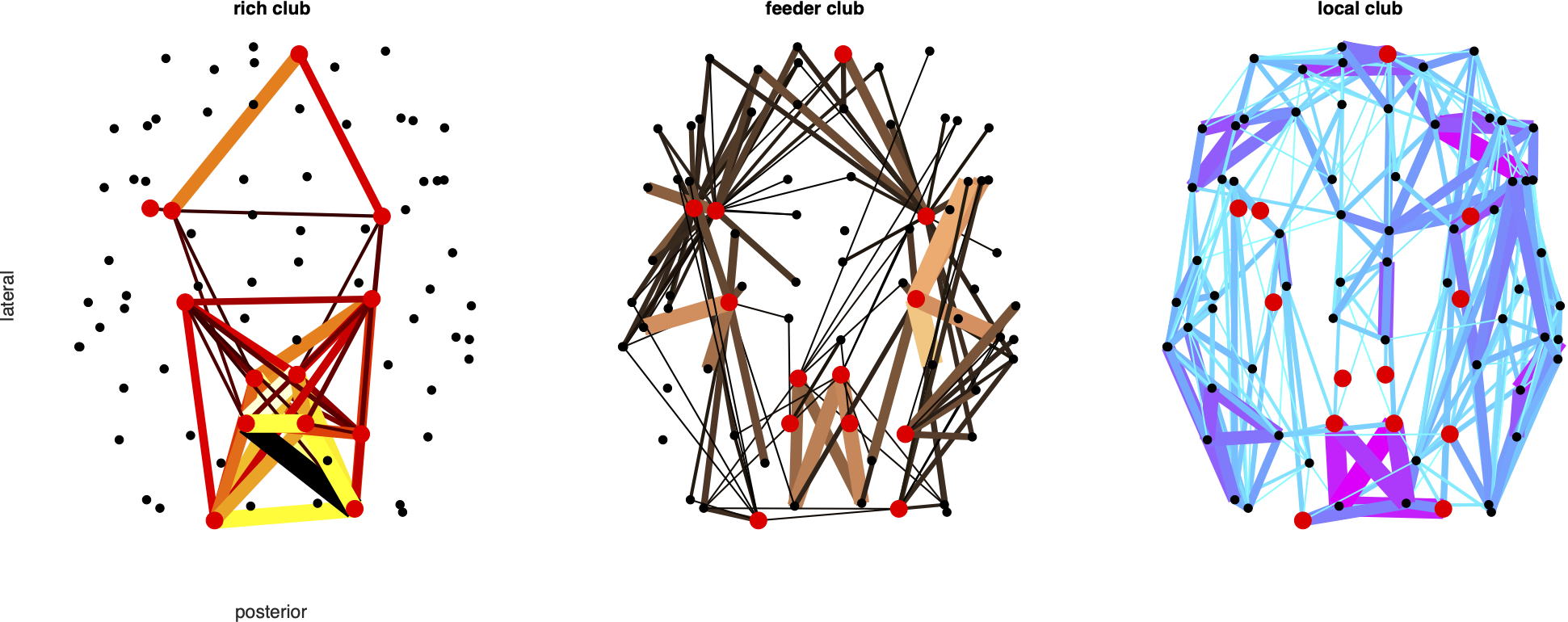
% C2. Rich clubs
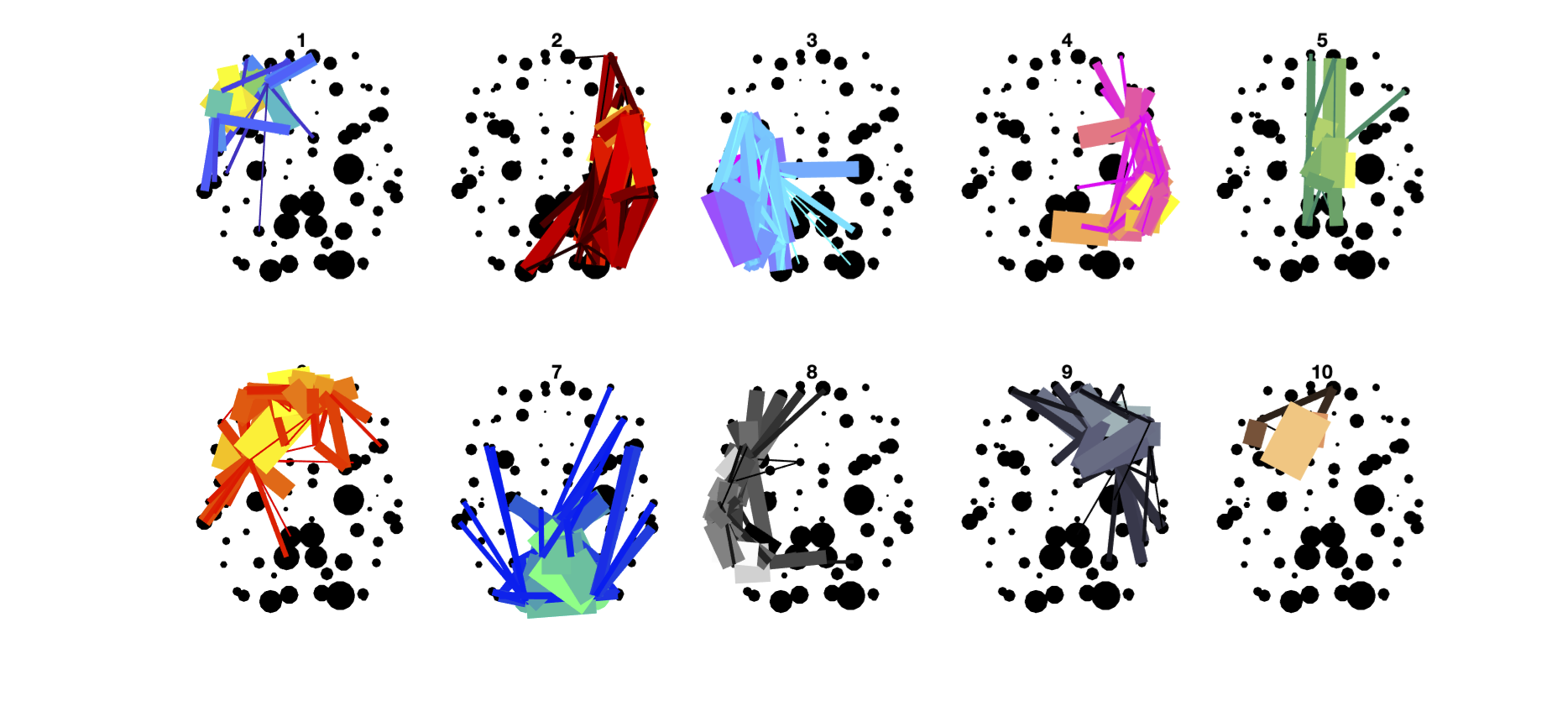
% C3. Edge categories
% C4. Percolation
%
% D: Binary [selected analyses]
%
% D1: Measures
% D2: Clustering
% D3: Path length
% D4: Symmetry
% D5: Hubs
% D6: Rich club
%
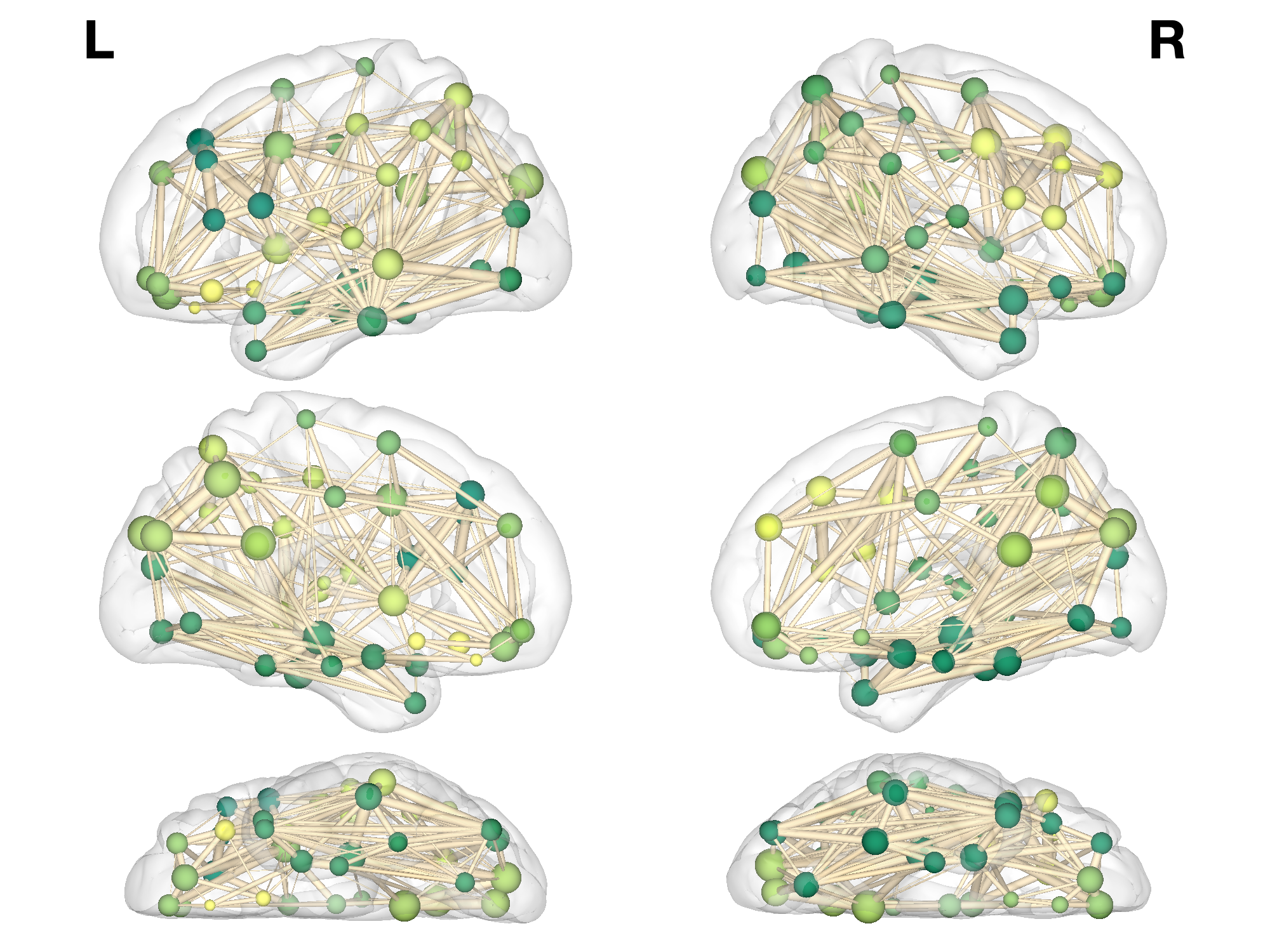
% E: Visualisation
%
% E1: Basic
% E2: Edge cost
% E3: Spheres
% E4: Growing
% E5: Rich club
% E6: Modules
% E7: 3D
% E8: Rotating
% E9: Gephi
% E10:Neuromarvl
% E11:Circular (Schemaball)
% E12:BrainNet
% Michael Hart, University of British Columbia, February 2021
%% A1. Load data
%This should be the only part required to be set manually
%Directory
directory = '/path_to_my_data';
%Patient ID
patientID = 'my_patient_ID';
%Template name
template = 'AAL90';